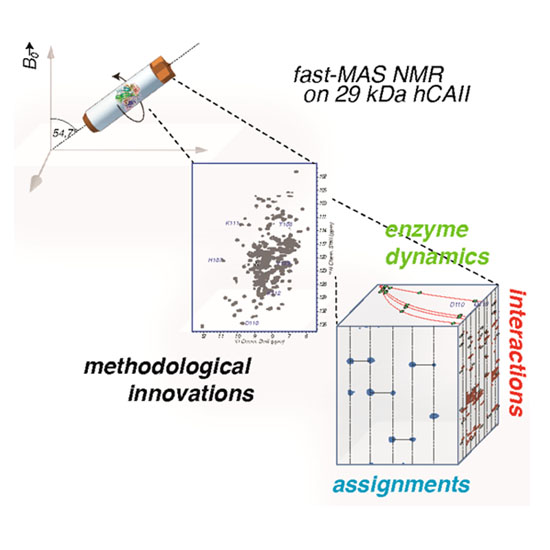
Dynamics and Interactions of a 29 kDa Human Enzyme Studied by Solid-State NMR
26-Feb-2018
J. Phys. Chem. Lett., 2018, 9 (6), pp 1307–1311, DOI: 10.1021/acs.jpclett.8b00110
J. Phys. Chem. Lett., online article
Solid-state NMR has been employed for characterization of a broad range of biomacromolecules and supramolecular assemblies. However, because of limitations in sensitivity and resolution, the size of the individual monomeric units has rarely exceeded 15 kDa. As such, enzymes, which are often more complex and comprise long peptide chains, have not been easily accessible, even though manifold desirable information could potentially be provided by solid-state NMR studies. Here, we demonstrate that more than 1200 backbone and side-chain chemical shifts can be reliably assessed from minimal sample quantities for a 29 kDa human enzyme of the carbonic anhydrase family, giving access to its backbone dynamics and intermolecular interactions with a small-molecule inhibitor. The possibility of comprehensive assessment of enzymes in this molecular-weight regime without molecular-tumbling-derived limitations enables the study of residue-specific properties important for their mode of action as well as for pharmacological interference in this and many other enzymes.