Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation
13-Jul-2014
Nature Structural & Molecular Biology, 2014, doi:10.1038/nsmb.2855, 21, 671–678 published on 13.07.2014
Nature Structural & Molecular Biology, online article
Nature Structural & Molecular Biology, online article
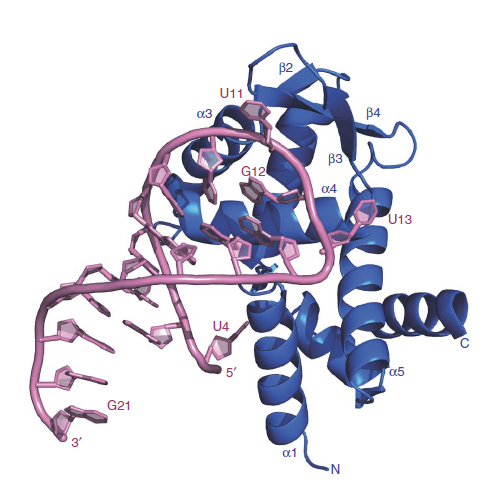
Roquin function in T cells is essential for the prevention of autoimmune disease. Roquin interacts with the 3′ untranslated regions (UTRs) of co-stimulatory receptors and controls T-cell activation and differentiation. Here we show that the N-terminal ROQ domain from mouse roquin adopts an extended winged-helix (WH) fold, which is sufficient for binding to the constitutive decay element (CDE) in the Tnf 3′ UTR. The crystal structure of the ROQ domain in complex with a prototypical CDE RNA stem-loop reveals tight recognition of the RNA stem and its triloop. Surprisingly, roquin uses mainly non-sequence-specific contacts to the RNA, thus suggesting a relaxed CDE consensus and implicating a broader spectrum of target mRNAs than previously anticipated. Consistently with this, NMR and binding experiments with CDE-like stem-loops together with cell-based assays confirm roquin-dependent regulation of relaxed CDE consensus motifs in natural 3′ UTRs.