Computational study of remodeling in a nucleosomal array
10-Aug-2015
The European Physical Journal E, 2015, DOI 10.1140/epje/i2015-15085-4, 38:85 published on 10.08.2015
The European Physical Journal E, online article
The European Physical Journal E, online article
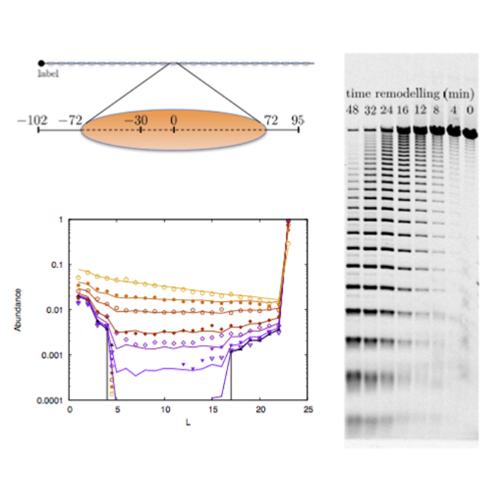
Chromatin remodeling complexes utilize the energy of ATP hydrolysis to change the packing state of chromatin, e.g. by catalysing the sliding of nucleosomes along DNA. Here we present simple models to describe experimental data of changes in DNA accessibility along a synthetic, repetitive array of nucleosomes during remodeling by the ACF enzyme or its isolated ATPase subunit, ISWI. We find substantial qualitative differences between the remodeling activities of ISWI and ACF. To understand better the observed behavior for the ACF remodeler, we study more microscopic models of nucleosomal arrays