Top-Down de Novo Protein Sequencing of a 13.6 kDa Camelid Single Heavy Chain Antibody by Matrix-Associated Laser Desorption Ionization-Time-of-Flight/Time-of-Flight Mass Spectrometry
23-Mar-2010
Analytical Chemistry, 2010, doi:10.1021/ac1000515, published on 23.03.2010
Analytical Chemistry, online article
Analytical Chemistry, online article
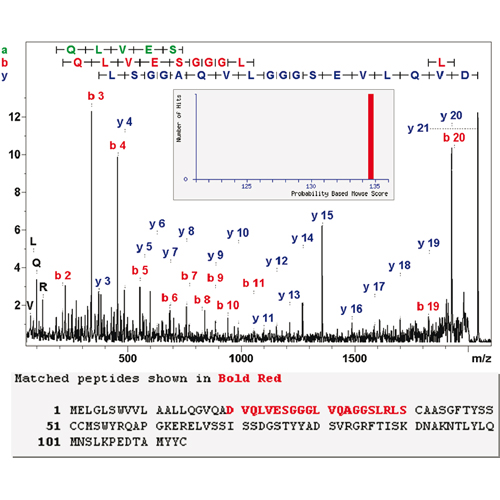
The primary structure of a 13.6 kDa single heavy chain camelid antibody (VHH) was determined by matrix-assisted laser desorption ionization-time-of-flight/time-of-flight (MALDI-TOF/TOF) top-down sequence analysis. The majority of the sequence was obtained by mass spectrometric de novo sequencing, with the N-terminal 14 amino acid residues being determined using T3-sequencing and database interrogation. The determined sequence was confirmed by liquid chromatography−tandem mass spectrometry (LC−MS/MS) analysis of a tryptic digest, which also provided high-energy collisionally induced dissociation (CID) data permitting the clear assignment of 3 of the 14 isobaric Leu/Ile residues. Five of the 11 Leu/Ile ambiguities could be resolved by homology comparisons with known VHH sequences. The monoisotopic molecular weight of the VHH was determined by ultrahigh-resolution orthogonal electrospray (ESI)-TOF analysis and found to be 13610.6066 Da, in excellent agreement with the established sequence. To our knowledge, this is the first time that the entire primary structure of a protein with a molecular weight >13 kDa has been established by mass spectrometric top-down sequencing.