Single-molecule force spectroscopy on polyproteins and receptor–ligand complexes: The current toolbox
09-Feb-2016
Journal of Structural Biology, DOI: http://dx.doi.org/10.1016/j.jsb.2016.02.011
Journal of Structural Biology, online article
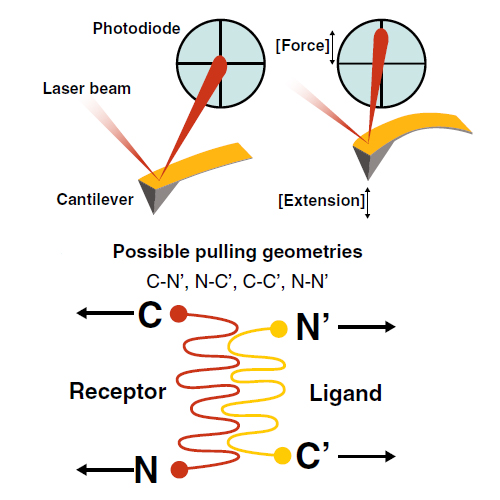
Single-molecule force spectroscopy sheds light onto the free energy landscapes governing protein folding and molecular recognition. Since only a single molecule or single molecular complex is probed at any given point in time, the technique is capable of identifying low-probability conformations within a large ensemble of possibilities. It furthermore allows choosing certain unbinding pathways through careful selection of the points at which the force acts on the protein or molecular complex. This review focuses on recent innovations in construct design, site-specific bioconjugation, measurement techniques, instrumental advances, and data analysis methods for improving workflow, throughput, and data yield of AFM-based single-molecule force spectroscopy experiments. Current trends that we highlight include customized fingerprint domains, peptide tags for site-specific covalent surface attachment, and polyproteins that are formed through mechanostable receptor–ligand interactions. Recent methods to improve measurement stability, signal-to-noise ratio, and force precision are presented, and theoretical considerations, analysis methods, and algorithms for analyzing large numbers of force–extension curves are further discussed. The various innovations identified here will serve as a starting point to researchers in the field looking for opportunities to push the limits of the technique further.