Biosynthesis of Isoprenoids: Crystal Structure of the [4Fe–4S] Cluster Protein IspG
10-Dec-2010
Journal of Molecular Biology,, 2010, doi:10.1016/j.jmb.2010.09.050, Volume 404, Issue 4, Pages 600-610 published on 10.12.2010
Journal of Molecular Biology, online article
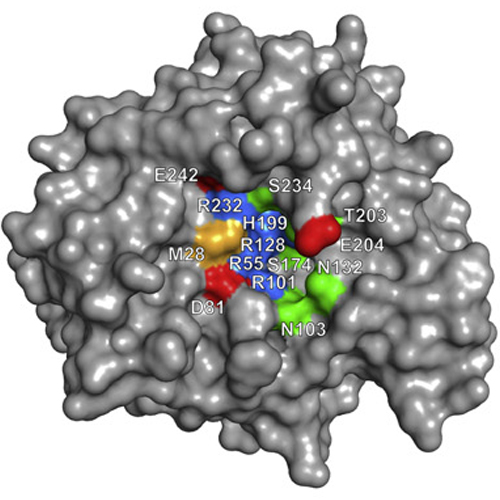
IspG protein serves as the penultimate enzyme of the recently discovered non-mevalonate pathway for the biosynthesis of the universal isoprenoid precursors, isopentenyl diphosphate and dimethylallyl diphosphate. The enzyme catalyzes the reductive ring opening of 2C-methyl-d-erythritol 2,4-cyclodiphosphate, which affords 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate. The protein was crystallized under anaerobic conditions, and its three-dimensional structure was determined to a resolution of 2.7 Å. Each subunit of the c2 symmetric homodimer folds into two domains connected by a short linker sequence. The N-terminal domain (N domain) is an eight-stranded β barrel that belongs to the large TIM-barrel superfamily. The C-terminal domain (C domain) consists of a β sheet that is flanked on both sides by helices. One glutamate and three cysteine residues of the C domain coordinate a [4Fe–4S] cluster. Homodimer formation involves an extended contact area (about 1100 Å2) between helices 8 and 9 of each respective β barrel. Moreover, each C domain contacts the N domain of the partner subunit, but the interface regions are small (about 430 Å2). We propose that the enzyme substrate binds to the positively charged surface area at the C-terminal pole of the β barrel. The C domain carrying the iron–sulfur cluster could then move over to form a closed conformation where the substrate is sandwiched between the N domain and the C domain. This article completes the set of three dimensional structures of the non-mevalonate pathway enzymes, which are of specific interest as potential targets for tuberculostatic and antimalarial drugs.