Crystal Structure and Mechanism of a DNA (6-4) Photolyase
27-Oct-2008
Angew. Chem. Int. Ed., 2008, 47, 10.1002/anie.200804268 published on 27.10.2008
Angewandte Chemie Int. Ed., online article
Angewandte Chemie Int. Ed., online article
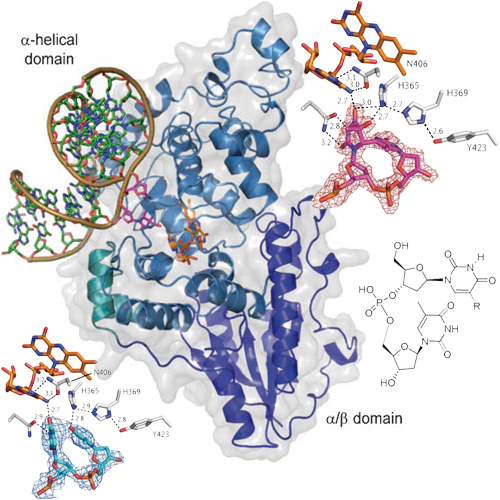
UV irradiation of cells gives rise to the formation of cyclobutane pyrimidine dimers (CPD) and so-called (6-4) DNA lesions (Scheme 1). Both lesions are major photoproducts formed in dipyrimidine sequences of double-stranded DNA. Repair of these lesions is essential because of their high mutagenic potential. Particularly important in many organisms are the photolyase-mediated repair systems that are able to split CPD lesions and (6-4) lesions directly back into their corresponding monomers. While formation and photolyase repair of CPD lesions is well studied, little is known about (6-4) lesions. In particular, the mechanism of repair of the (6-4) lesions by (6-4) DNAphotolyases is a longstanding question. Currently it is believed that the enzyme rearranges the (6-4) lesion with the help of two conserved histidine residues in the active site to form an oxetane intermediate (Scheme 1), which is split after single-electron donation from a light-activated FADH. We report here the first crystal structures of a (6-4) DNA photolyase enzyme. The structures show the enzyme in complex with a (6-4) lesion containing DNAbefore and after in situ repair. Based on the structural and biochemical data we propose a modified repair mechanism that lacks the strained oxetane intermediate.